Learning Objectives
There are two broad categories of Supervised Classification based on the type of response your modeling \(y \sim x\), where \(y\) is either a continuous value, in which case Regression, or it is a categorical value, in which case it’s a Classification.
A binary response, such as presence or absence, is a categorical value, so typically a Classification technique would be used. However, by transforming the response with a logit function, we were able to use Regression techniques like generalized linear (glm()) and generalized additive (gam()) models.
In this portion of the lab you’ll use Decision Trees as a Classification technique to the data with the response being categorical (factor(present)).
Recursive Partitioning (
rpart())
Originally called classification & regression trees (CART), but that’s copyrighted (Breiman, 1984).Random Forest (
ranger())
Actually an ensemble model, ie trees of trees.
1 Setup
# global knitr chunk options
knitr::opts_chunk$set(
warning = FALSE,
message = FALSE)
# load packages
librarian::shelf(
caret, # m: modeling framework
dplyr, ggplot2 ,here, readr,
pdp, # X: partial dependence plots
ranger, # m: random forest modeling
rpart, # m: recursive partition modeling
rpart.plot, # m: recursive partition plotting
rsample, # d: split train/test data
skimr, # d: skim summarize data table
vip) # X: variable importance
# options
options(
scipen = 999,
readr.show_col_types = F)
set.seed(42)
# graphical theme
ggplot2::theme_set(ggplot2::theme_light())
# paths
dir_data <- here("data/sdm")
pts_env_csv <- file.path(dir_data, "pts_env.csv")
# read data
pts_env <- read_csv(pts_env_csv)
d <- pts_env %>%
select(-ID) %>% # not used as a predictor x
mutate(
present = factor(present)) %>% # categorical response
na.omit() # drop rows with NA
skim(d)
| Name | d |
| Number of rows | 986 |
| Number of columns | 8 |
| _______________________ | |
| Column type frequency: | |
| factor | 1 |
| numeric | 7 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| present | 0 | 1 | FALSE | 2 | 0: 499, 1: 487 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| WC_alt | 0 | 1 | 341.77 | 422.02 | 3.00 | 83.00 | 204.50 | 461.75 | 3999.00 | ▇▁▁▁▁ |
| WC_bio1 | 0 | 1 | 24.72 | 2.60 | 4.70 | 23.80 | 25.60 | 26.40 | 28.50 | ▁▁▁▂▇ |
| WC_bio2 | 0 | 1 | 10.07 | 1.67 | 6.10 | 8.92 | 9.80 | 11.10 | 15.40 | ▂▇▆▃▁ |
| ER_tri | 0 | 1 | 30.76 | 35.72 | 0.47 | 6.25 | 14.61 | 41.50 | 223.22 | ▇▂▁▁▁ |
| ER_topoWet | 0 | 1 | 11.16 | 1.63 | 7.14 | 10.07 | 11.47 | 12.47 | 14.69 | ▂▃▆▇▂ |
| lon | 0 | 1 | -66.40 | 14.72 | -86.06 | -79.71 | -67.62 | -54.15 | -34.84 | ▇▅▃▅▂ |
| lat | 0 | 1 | -1.36 | 10.50 | -23.71 | -9.52 | -0.50 | 9.12 | 13.54 | ▂▃▅▃▇ |
1.1 Split data into training and testing
# create training set with 80% of full data
d_split <- rsample::initial_split(d, prop = 0.8, strata = "present")
d_train <- rsample::training(d_split)
# show number of rows present is 0 vs 1
table(d$present)
0 1
499 487 table(d_train$present)
0 1
399 389 2 Decision Trees
Reading: Boehmke & Greenwell (2020) Hands-On Machine Learning with R. Chapter 9 Decision Trees
2.1 Partition, depth=1
# run decision stump model
mdl <- rpart(
present ~ ., data = d_train,
control = list(
cp = 0, minbucket = 5, maxdepth = 1))
mdl
n= 788
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 788 389 0 (0.50634518 0.49365482)
2) lat< 7.7401 512 128 0 (0.75000000 0.25000000) *
3) lat>=7.7401 276 15 1 (0.05434783 0.94565217) *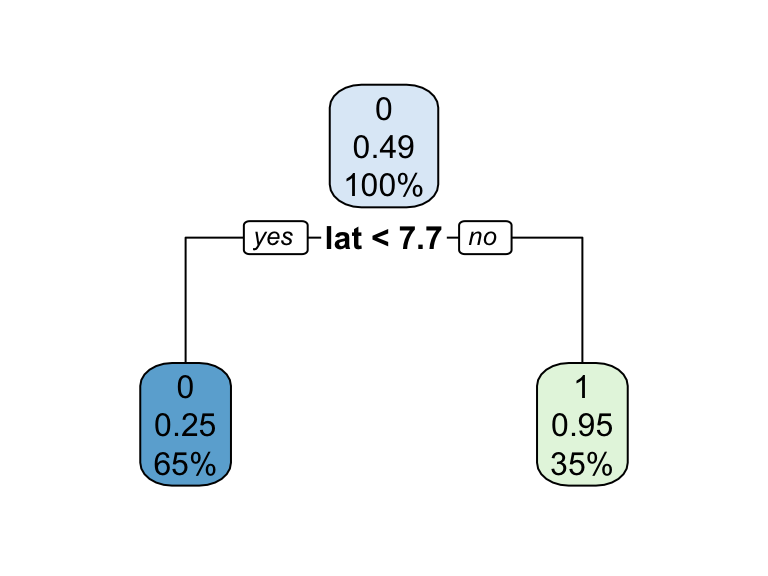
Figure 1: Decision tree illustrating the single split on feature x (left).
2.2 Partition, depth=default
# decision tree with defaults
mdl <- rpart(present ~ ., data = d_train)
mdl
n= 788
node), split, n, loss, yval, (yprob)
* denotes terminal node
1) root 788 389 0 (0.50634518 0.49365482)
2) lat< 7.7401 512 128 0 (0.75000000 0.25000000)
4) WC_bio2>=10.25 247 22 0 (0.91093117 0.08906883) *
5) WC_bio2< 10.25 265 106 0 (0.60000000 0.40000000)
10) WC_bio1>=26.15 87 10 0 (0.88505747 0.11494253) *
11) WC_bio1< 26.15 178 82 1 (0.46067416 0.53932584)
22) lat>=0.617405 32 5 0 (0.84375000 0.15625000) *
23) lat< 0.617405 146 55 1 (0.37671233 0.62328767)
46) lat< -2.529476 77 36 0 (0.53246753 0.46753247)
92) lat>=-16.94087 45 8 0 (0.82222222 0.17777778)
184) WC_bio2>=7.2 38 2 0 (0.94736842 0.05263158) *
185) WC_bio2< 7.2 7 1 1 (0.14285714 0.85714286) *
93) lat< -16.94087 32 4 1 (0.12500000 0.87500000) *
47) lat>=-2.529476 69 14 1 (0.20289855 0.79710145)
94) lon>=-75.44386 33 12 1 (0.36363636 0.63636364)
188) WC_alt>=67 12 0 0 (1.00000000 0.00000000) *
189) WC_alt< 67 21 0 1 (0.00000000 1.00000000) *
95) lon< -75.44386 36 2 1 (0.05555556 0.94444444) *
3) lat>=7.7401 276 15 1 (0.05434783 0.94565217) *rpart.plot(mdl)
# plot complexity parameter
plotcp(mdl)
# rpart cross validation results
mdl$cptable
CP nsplit rel error xerror xstd
1 0.63239075 0 1.0000000 1.1028278 0.03593877
2 0.03084833 1 0.3676093 0.3958869 0.02861490
3 0.01542416 6 0.2005141 0.2699229 0.02452406
4 0.01285347 8 0.1696658 0.2622108 0.02422420
5 0.01000000 9 0.1568123 0.2596401 0.02412272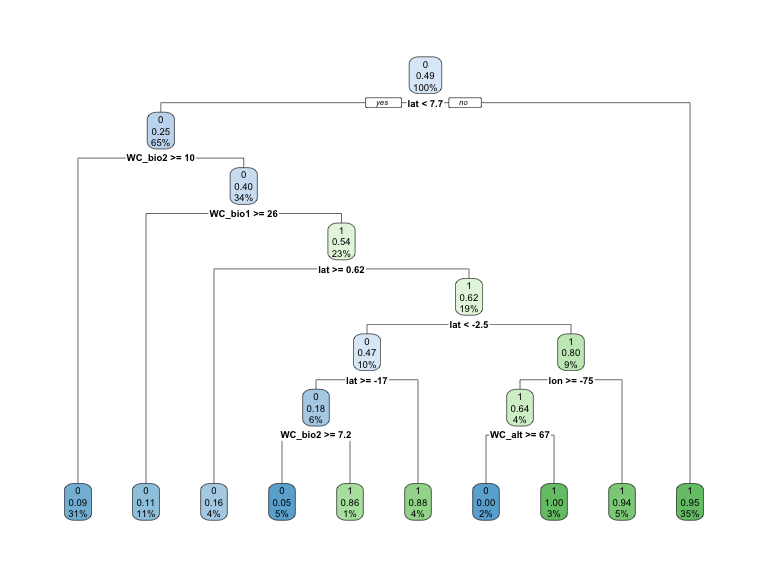
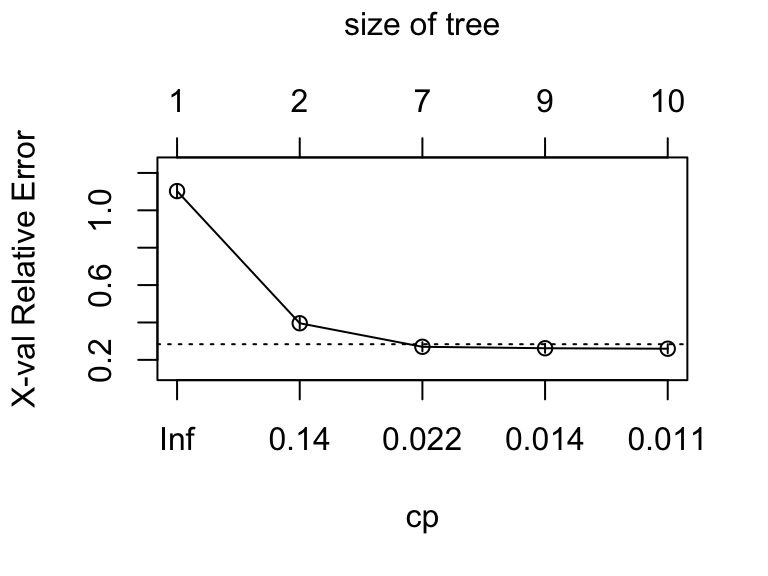
Figure 2: Decision tree \(present\) classification.
2.3 Feature interpretation
# caret cross validation results
mdl_caret <- train(
present ~ .,
data = d_train,
method = "rpart",
trControl = trainControl(method = "cv", number = 10),
tuneLength = 20)
ggplot(mdl_caret)
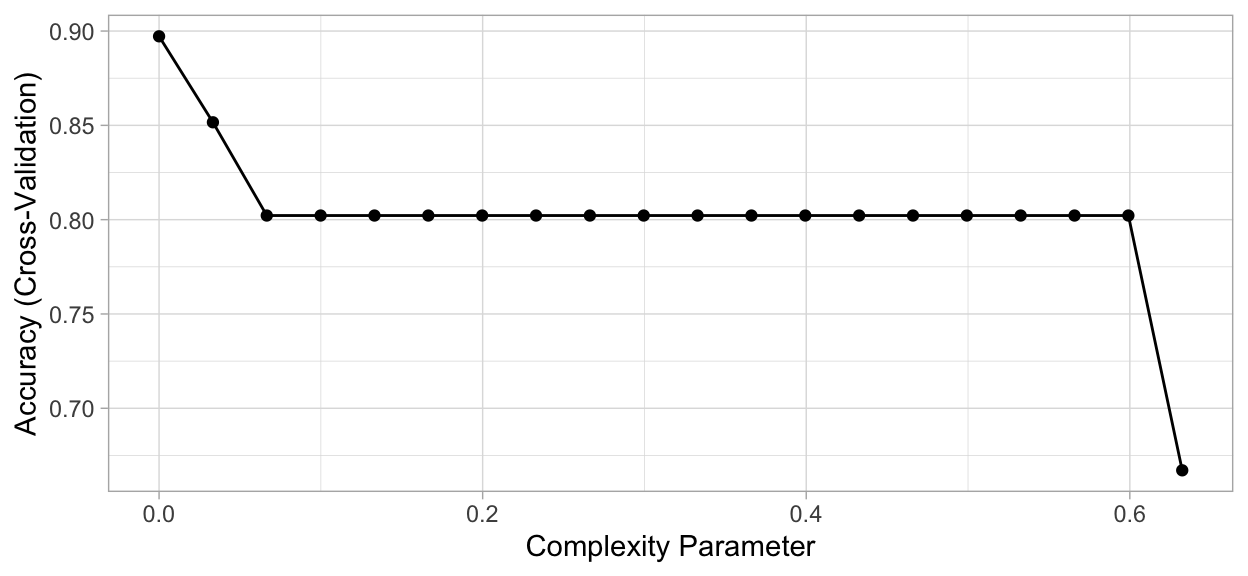
Figure 3: Cross-validated accuracy rate for the 20 different \(\alpha\) parameter values in our grid search. Lower \(\alpha\) values (deeper trees) help to minimize errors.
vip(mdl_caret, num_features = 40, bar = FALSE)
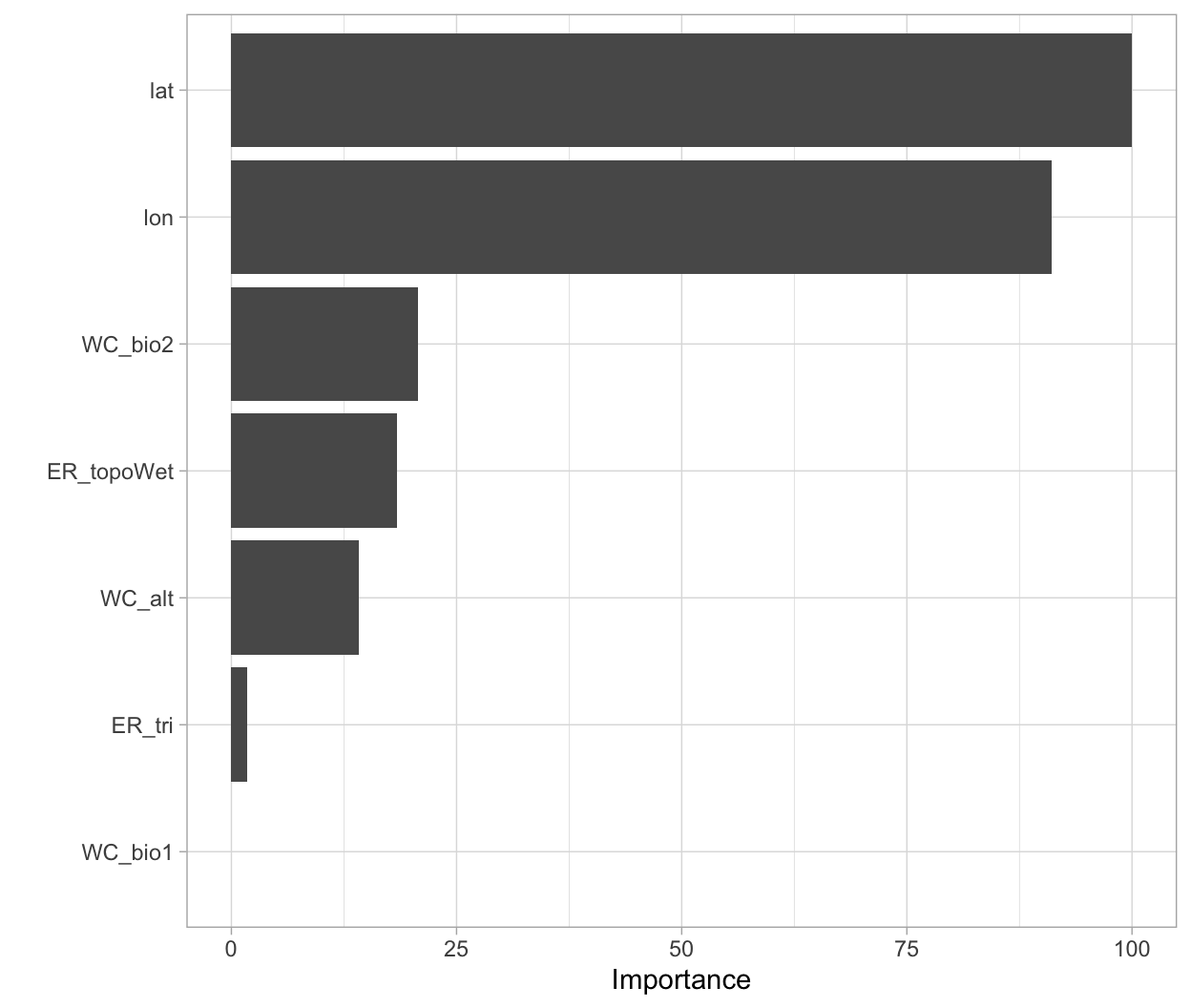
Figure 4: Variable importance based on the total reduction in MSE for the Ames Housing decision tree.
# Construct partial dependence plots
p1 <- partial(mdl_caret, pred.var = "lat") %>% autoplot()
p2 <- partial(mdl_caret, pred.var = "WC_bio2") %>% autoplot()
p3 <- partial(mdl_caret, pred.var = c("lat", "WC_bio2")) %>%
plotPartial(levelplot = FALSE, zlab = "yhat", drape = TRUE,
colorkey = TRUE, screen = list(z = -20, x = -60))
# Display plots side by side
gridExtra::grid.arrange(p1, p2, p3, ncol = 3)
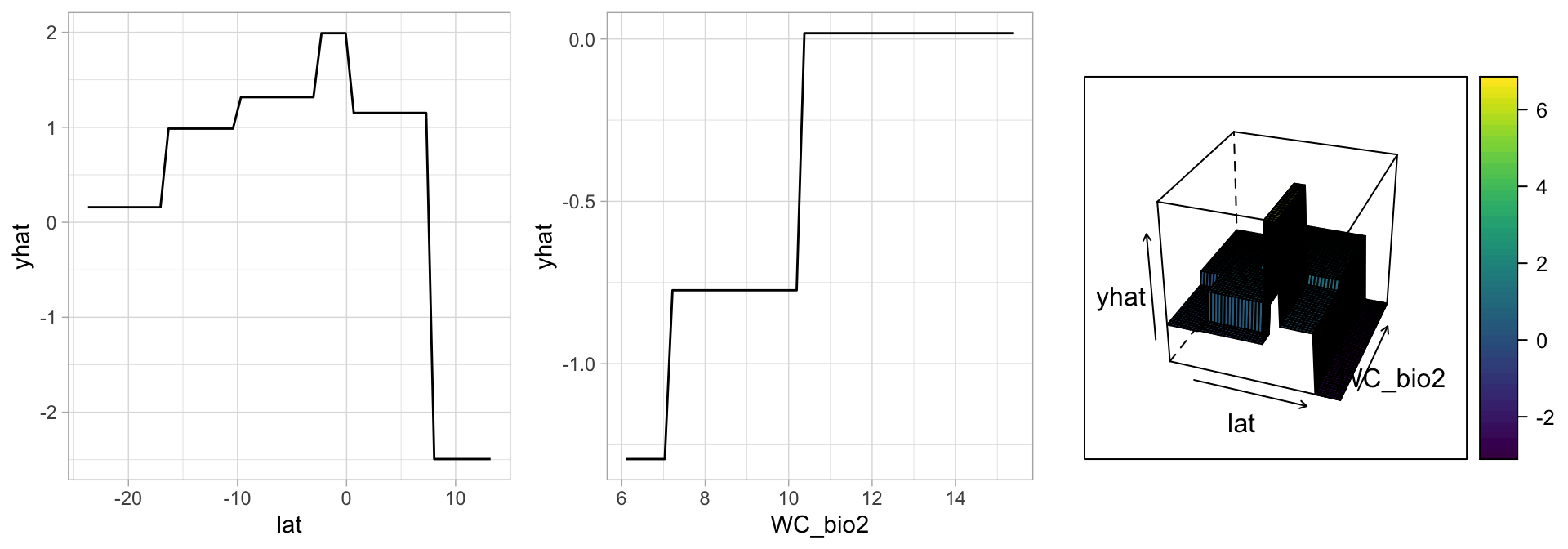
Figure 5: Partial dependence plots to understand the relationship between lat, WC_bio2 and present.
3 Random Forests
Reading: Boehmke & Greenwell (2020) Hands-On Machine Learning with R. Chapter 11 Random Forests
See also: Random Forest – Modeling methods | R Spatial
3.1 Fit
# number of features
n_features <- length(setdiff(names(d_train), "present"))
# fit a default random forest model
mdl_rf <- ranger(present ~ ., data = d_train)
# get out of the box RMSE
(default_rmse <- sqrt(mdl_rf$prediction.error))
[1] 0.25189643.2 Feature interpretation
# re-run model with impurity-based variable importance
mdl_impurity <- ranger(
present ~ ., data = d_train,
importance = "impurity")
# re-run model with permutation-based variable importance
mdl_permutation <- ranger(
present ~ ., data = d_train,
importance = "permutation")
p1 <- vip::vip(mdl_impurity, bar = FALSE)
p2 <- vip::vip(mdl_permutation, bar = FALSE)
gridExtra::grid.arrange(p1, p2, nrow = 1)
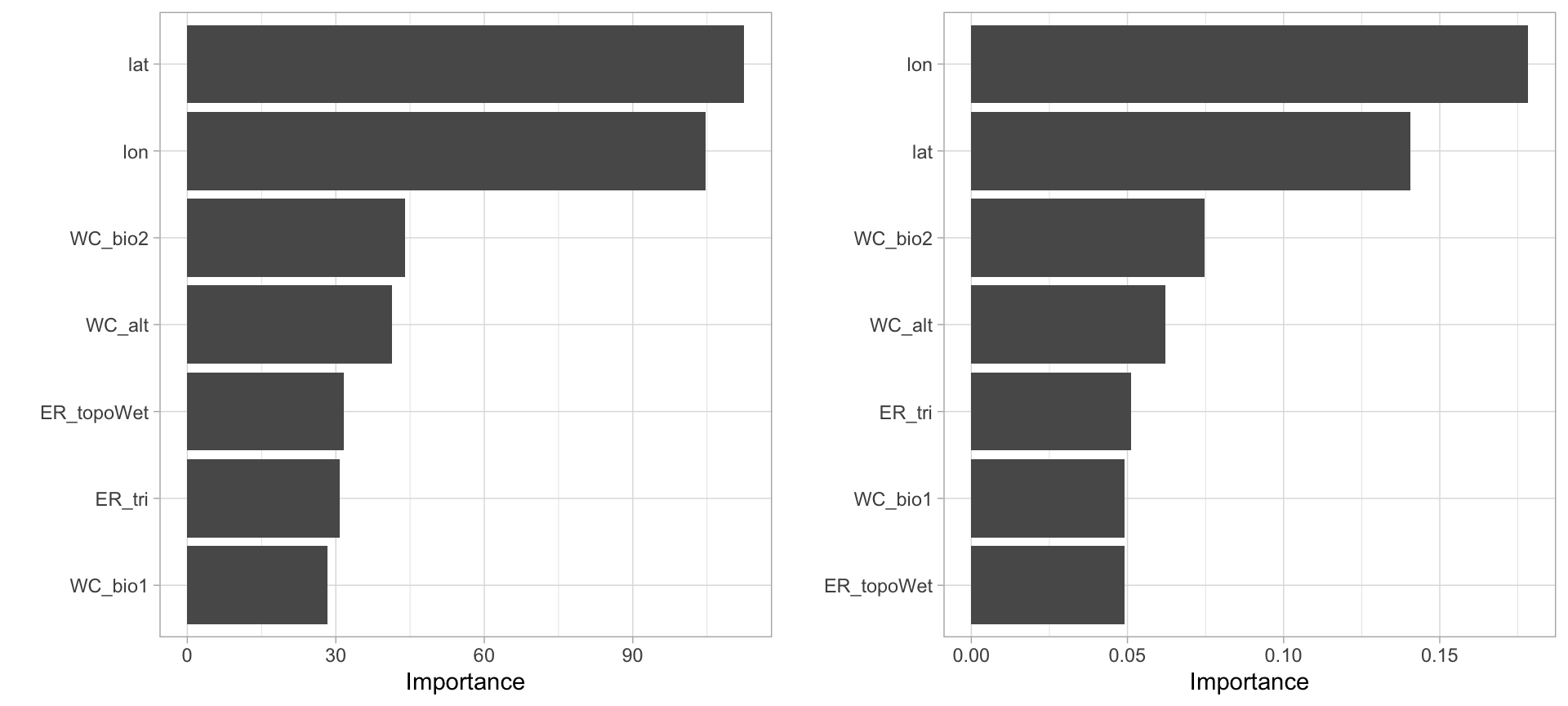
Figure 6: Most important variables based on impurity (left) and permutation (right).
